Themes

Context
On the other hand, physicians also describe and write down important patient characteristics and disease-related symptoms for almost every medical encounter through free-form medical notes. In recent years, significant advances in multi-omics technology (e.g., genomics, transcriptomics, etc.) have also created unprecedented opportunities to characterize biological processes correlated with disease. However, combining these heterogeneous data sources in a meaningful way for disease prediction is challenging.
Therefore, for better precision medicine, physicians must now make increasingly complex therapeutic decisions with an unrealistic number of variables. Therefore, developments in artificial intelligence (AI) are envisioned to create a data science revolution in medicine. In particular, graphical neural networks (GNNs) have shown immense potential in learning meaningful and powerful data representations by combining the relational inference of graphical models with the power of deep learning. However, since the power of deep learning is strongly associated with data size and medical data cannot be easily shared among medical institutions due to patient privacy concerns, developing powerful GNN models for disease prediction in healthcare is a major challenge.
Mission
- (i) GNN models can be developed from the databases of multiple healthcare facilities, thereby increasing the size of the analyzed data; and
- (ii) data are always kept within the boundaries of each healthcare facility, thus avoiding data transfer.
The figures below provide an overview of the main research framework of the MEDomics UdeS lab.

Research Area 1: Medical Imaging
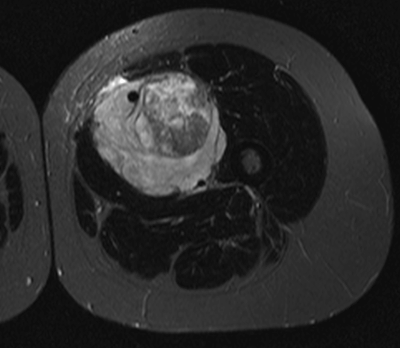
The most aggressive tumors tend to be more heterogeneous
Neural networks can detect the most heterogeneous tumors
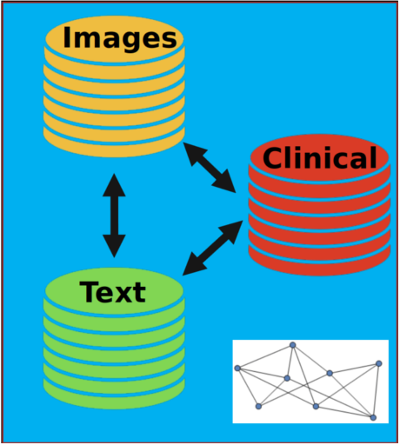
Research Area 2: Heterogeneous Data
Data from different sources can be combined to make better predictions
For example, medical imaging, physician text notes and clinical data are full of relevant information
Research Area 3: Federated Learning
To guarantee patient privacy while ensuring maximum use of relevant data
Decentralized learning to ensure medical center sovereignty
MEDomicsTools
The MEDomics UdeS lab aims to promote the use of best practices in data science.

This laboratory is a sub-branch of the MEDomics consortium.
Visit the MEDomics website Main publication of the consortium

